
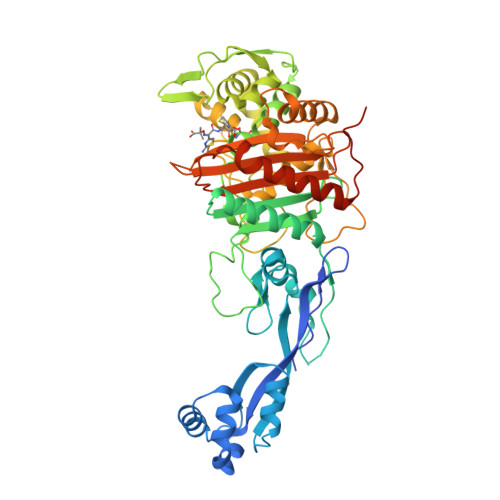
We were successful in the generation of a S. Altogether, these results indicated that PBP3 SAL evolved for promoting cell division in acidic environments.Īdditional experiments sustained a role of PBP3 SAL as an enzyme promoting cell division during the interaction of S. coli strain producing a thermosensitive PBP3 only when bacteria were grown in acidified medium. This observation agreed with the fact that PBP3 SAL restored cell division at 42☌ in an E. Interestingly, our initial experiments showed that, unlike PBP3, the PBP3 SAL was able to bind Boc-FL “exclusively” at acidic pH. Binding assays based on fluorescent β‑lactam antibiotics (Boc-FL) demonstrated that PBP3 SAL is a genuine PBP. This PBP3 paralogue, which we named PBP3 SAL, is also a class B PBP with a transpeptidase (TP) domain in which the three catalytic motifs, SXXK (containing the active serine site), SXN/D and KTG, are conserved. Typhimurium genome has a gene encoding a PBP3 paralogue (63% identity at the amino acid level). Our comparative genome analyses, directed to identify Salmonella-specific genes encoding new PG enzymes, revealed that, besides the ftsI gene encoding the PBP3 involved in cell division, the S. Typhimurium to minimize NOD1 recognition. The activity of this D-L-endopeptidase could be therefore exploited by intracellular S. This D-Glu- mDap motif is absolutely essential for NOD1 recognition. Typhimurium of an PG enzyme with D-L-endopeptidase activity that is absent in non-pathogenic bacteria and cleaves the D-glutamic acid(D-Glu)- meso-diamopimelic acid ( mDap) bond of the PG stem peptide. We reported recently the induction in intracellular S. Members of this defence mechanism include the PG-sensing receptors NOD1 and NOD2. Altered PG structure linked to the colonization of the intracellular niche has important implications for host defences that evolved to recognize PG fragments in an environment, normally not colonized by bacteria, e.g. Typhimurium utilizes distinct machinery for synthesis and remodelling of the cell envelope in these two different lifestyles (extra- versus intracellular) is mostly unknown, especially for its main component, the peptidoglycan (PG). Typhimurium exhibits lower growth rate than in the cytosol or the extracellular milieu. When adapted to the intra-phagosomal lifestyle, S. These acidic compartments are strikingly different compared to the nutrient media in which bacteria are usually grown in the laboratory, which are buffered to neutral pH to favour optimal growth. Although recent studies provided evidence for the capacity of this pathogen gaining access to the cytosol of certain host cell types, in most cases the intracellular bacteria reside within acidic phagosomes. Typhimurium) is an intracellular bacteria pathogen that invades and proliferates inside phagocytic and non-phagocytic eukaryotic cells. Salmonella enterica serovar Typhimurium ( S. Typhimurium mutant defective in PBP3, which cannot divide at neutral pH. Our work also revealed that it is possible to generate a S. Typhimurium, displays PG biosynthetic activity restricted to acidic conditions. The second enzyme, named PBP3 SAL, is absent in non-pathogenic bacteria and, at least in S. One of these two division PG enzymes is orthologue of the division-specific PBP3 of Escherichia coli. Our recent results obtained in the intracellular bacterial pathogen Salmonella enterica serovar Typhimurium challenges this view since this bacterium has two PBPs that can independently build the division septum. To date, this division‑specific PBP was reported as unique in all known bacteria and, as a consequence, “essential”. An exemption is the PBP dedicated to build the septal PG required to separate daughter cells during cell division. This dispensability has led to the widely accepted concept of functional redundancy for many PBPs. On an average, bacterial genomes harbour a minimum of 10 PBP-encoding genes, most of them non-essential. TP reactions, involving cleavage of the terminal D‑Ala-D-Ala bond in the stem peptide, are carried out by enzymes known generically as penicillin-binding proteins (PBPs) due to their capacity to bind β‑lactam antibiotics, which are D‑Ala-D-Ala structural analogues. The building subunits -muropeptides- are incorporated into the growing macromolecule by transglycolyslation (TG) and transpeptidation (TP) reactions, which constitute the last biosynthetic steps. The PG is synthesized by a stepwise process that includes cytosolic and periplasmic reactions. The major component of the cell wall is the peptidoglycan (PG) a giant macromolecule formed by glycan chains cross-linked by short peptides. The bacterial cell wall preserves cell integrity in response to external insults and the internal turgor pressure.


 0 kommentar(er)
0 kommentar(er)
